PathIntegrate
PathIntegrate Python package for pathway-based multi-omics data integration
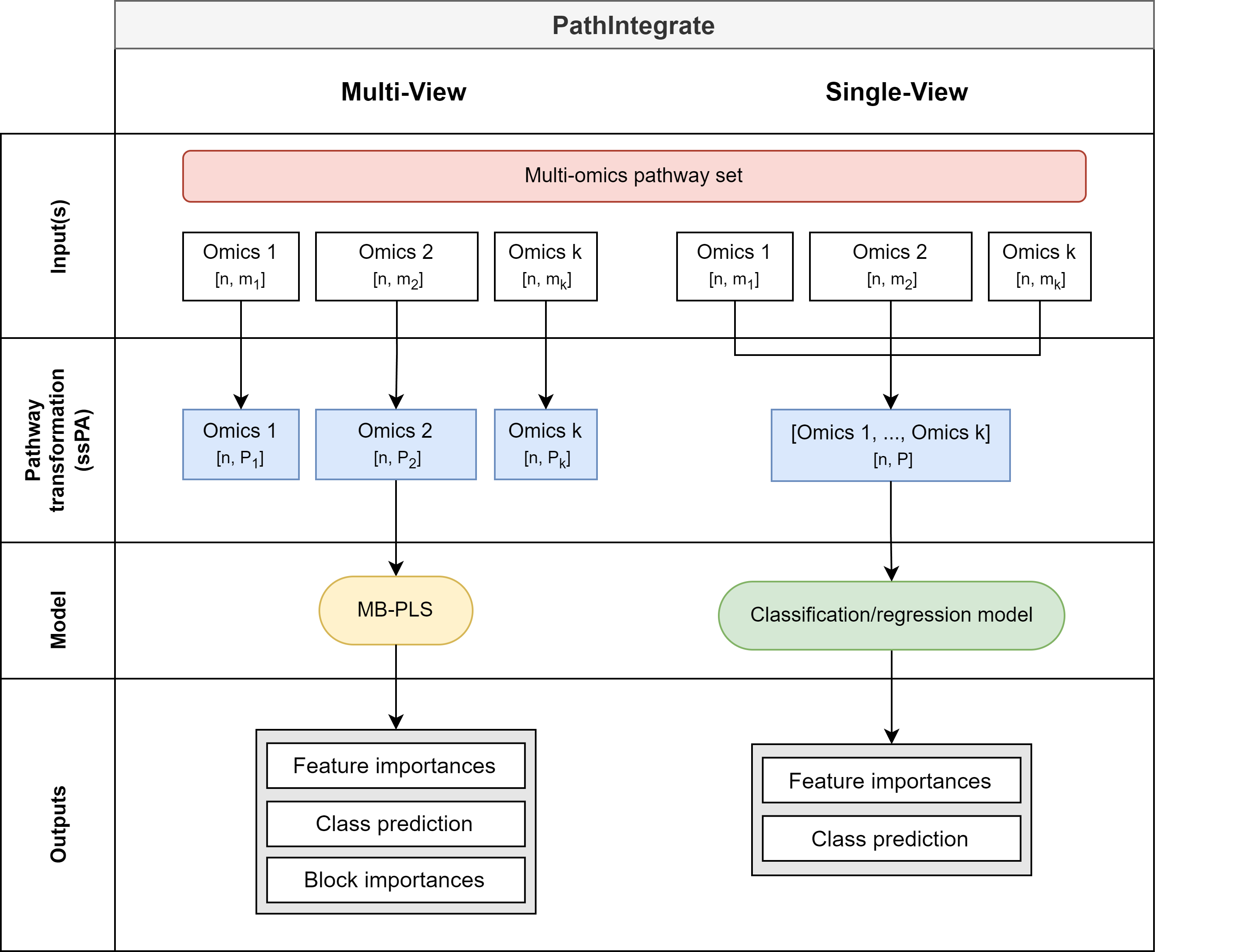
Features
- Pathway-based multi-omics data integration using PathIntegrate Multi-View and Single-View models
- Multi-View model: Integrates multiple omics datasets using a shared pathway-based latent space
- Single-View model: Integrates multi-omics data into one set of multi-omics pathway scores and applies an SKlearn-compatible predictive model
- Pathway importance
- Sample prediction
- SKlearn-like API for easy integration into existing pipelines
- Support for multiple pathway databases, including KEGG and Reactome
- Support for multiple pathway scoring methods available via the sspa package
- Cytoscape Network Viewer app for visualizing pathway-based multi-omics data integration results

Installation
pip install -i https://test.pypi.org/simple/ PathIntegrate
Tutorials and documentation
Please see our Quickstart guide on Google Colab
Citing PathIntegrate
If you use PathIntegrate in your research, please cite our paper:
PathIntegrate: Multivariate modelling approaches for pathway-based multi-omics data integration
Cecilia Wieder, Juliette Cooke, Clement Frainay, Nathalie Poupin, Jacob G. Bundy, Russell Bowler, Fabien Jourdan, Katerina J. Kechris, Rachel PJ Lai, Timothy Ebbels
Manuscript in preparation